A parallel version of Expo2014 is available if you want to speed up your crystal structure solution by direct space method (DSM) using more than one processor and exploiting the whole available computing power. Different parallelization paradigms are available to benefit from parallelism but in Expo2014 only Message Passing Interface (MPI) is at moment implemented and only DSM have been parallelized.
Running DSM in parallel permits tackling structures with great complexity in a reasonable time. Structures with more than 15-20 degrees of freedom, several molecular fragments, presence of largely flexible atomic chains represent a complex problem for the crystal structure solution by DSM: large number of SA moves per run, large number of runs are required to guarantee finding the global minimum and increasing the frequency of correct solutions. In addition, the use of dynamical occupancy correction in the case of non-molecular compounds or the introduction of constraints and restrains, requires the computation of geometrical parameters (distances and angles) between neighboring atoms including all symmetry equivalent positions. These expensive calculations could increase dramatically the computation time when there is a large number of atoms.
Fortunately the type of calculation for DSM can be easily distributed between more CPU-core. In a MPI job, different program copies are created, each copy has the own private memory and execute the same code independently from others but with the possibility to communicate with other by set of routines that make up the MPI library. In the parallel Expo2014 all the global optimization trials are distributed between the CPU-cores and each CPU-core runs a proper independent trials of structure solution. At the end the solutions found by the CPU-cores are communicated to the master process that orders all the structural models based on the cost function and performs output operations. In the current version of the program the CPU-cores do not exchange any data during the simulation or, in other words, the CPU-cores do not collaborate with each other. Because the overhead due to communications is practically zero, the parallel speedup (![]() ) is almost identical to the number of CPU-cores (
) is almost identical to the number of CPU-cores (![]() ). Implementations of the algorithm with periodical cooperation by exchanging the best solutions found between CPU-cores are under testing.
). Implementations of the algorithm with periodical cooperation by exchanging the best solutions found between CPU-cores are under testing.
Running the parallel version of EXPO requires:
- Computer with multi-core CPUs: laptop, desktop PC, workstation, supercomputer.
- Linux operating system installed.
- Open MPI installed.
- Install EXPO from source and linking with MPI libraries or from debian package.
- Run EXPO by using the launcher mpirun with the appropriate options.
Installing the parallel version of EXPO
The debian binary packages (see the download page) contain EXPO compiled with Open MPI. Alternatively, you can compile EXPO from source and link with MPI libraries. Read and follow the instructions in the section dedicated to the installation of EXPO on Linux platforms.
To use Expo in parallel mode in a Windows environment, you can use the Windows Subsystem for Linux (WSL). You can find the installation guide for WSL at the following link: https://docs.microsoft.com/en-us/windows/wsl/install.
The typical way to run Expo2014 under MPI is by using mpirun that is invoked from the command line with the arguments consisting of the number of MPI processes, the name of the program executable followed by the name of the input file, e.g.
mpirun -np 10 expo input_file.exp
Here the option -np specifies the number of MPI processes to run. The MPI process are the instances of program that are running and should be not more than the total number of CPU-cores to avoid degradation of performance. 10 processes are used in this case. The calculation will be distributed between 10 CPU-cores in a parallel computer with 10 or more CPU-cores. During a default DSM 10 run of the global optimization algorithm are performed so each CPU-core will perform 1 run. The number of runs can be increased by using the directive nrun in the input file. If the number of processes required exceeds the number of runs, the number of runs will be automatically extended to the number of processes exploiting all the hardware resources specified by the user with the np option.
The graphical user interface (GUI) is not displayed when Expo2014 runs in parallel, in order to monitor the progress of job, the user can consult the .out output file. When the calculation is completed, the program creates a project file structure_name.expo that can be used to display the results by GUI. In order to examine the results of the job, the .expo project file can be opened using Expo2014 from File > Old Project or typing the following command:
expo structure_name.expo
The best solution will be displayed. To access to the list of all structural models generated during the DSM select Solve > Simulated Annealing and click on the ![]() button.
button.
You can test the parallel version of Expo2014 on the structure paracetamol reported in the examples directory. Copy the file paracetamol.exp, paracetamol.xy and paracetamol.mol in a directory whose name might be test_mpi, change the working direcory in test_mpi.
mkdir test_mpi cd test_mpi cp /usr/local/share/expo/examples/paracetamol.* .
In the working directory test_mpi run the following command using a PC-biprocessor machine:
mpirun -np 2 expo paracetamol
where instead of expo you might declare the complete path of the folder where Expo2014 parallel is installed. In few minutes you should get something like this picture
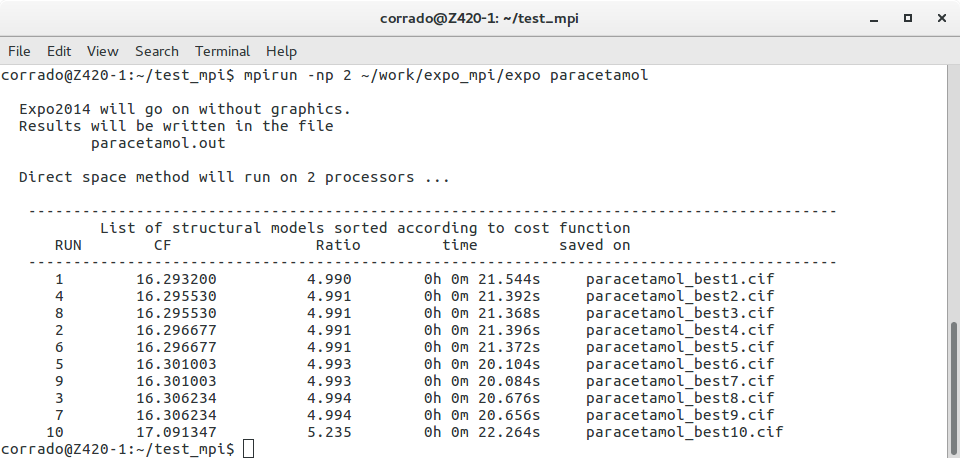
If you look the content of the working directory test_mpi (use command ls), some output files have been created: paracetamol.out containing general information about the job, CIF files paracetamol_best1.cif, paracetamol_best2.cif, … with the best structural models, a project file called paracetamol.expo. Load paracetamol.expo from File > Old Project or by the following command:
expo paracetamol.expo
Then Solve > Simulated Annealing and click on the ![]() button for a visual inspection of the best solutions.
button for a visual inspection of the best solutions.