The command %refine is used to improve phases produced by a previous run of Sir2019 or by a different program.
It is possible to create the input file for Sir2019 using a text editor; as an example:
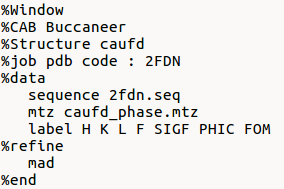
The full list of directives is available at the end of this page or here. The directives for %data, in case of macromolecules, are available here
The user may be interested to refine by Sir2019 phases obtained by other programs: e.g., phases obtained by a previous application of MR tool, or obtained as output of some SAD-MAD program, or otherwise previously refined. The instructions in this sections, valid for macromolecules only, help to perform the refinement task.
A graphic interface is available in Sir2019 to refine phases by means of the New item ![]() in the toolbar or in the menu File :
in the toolbar or in the menu File :
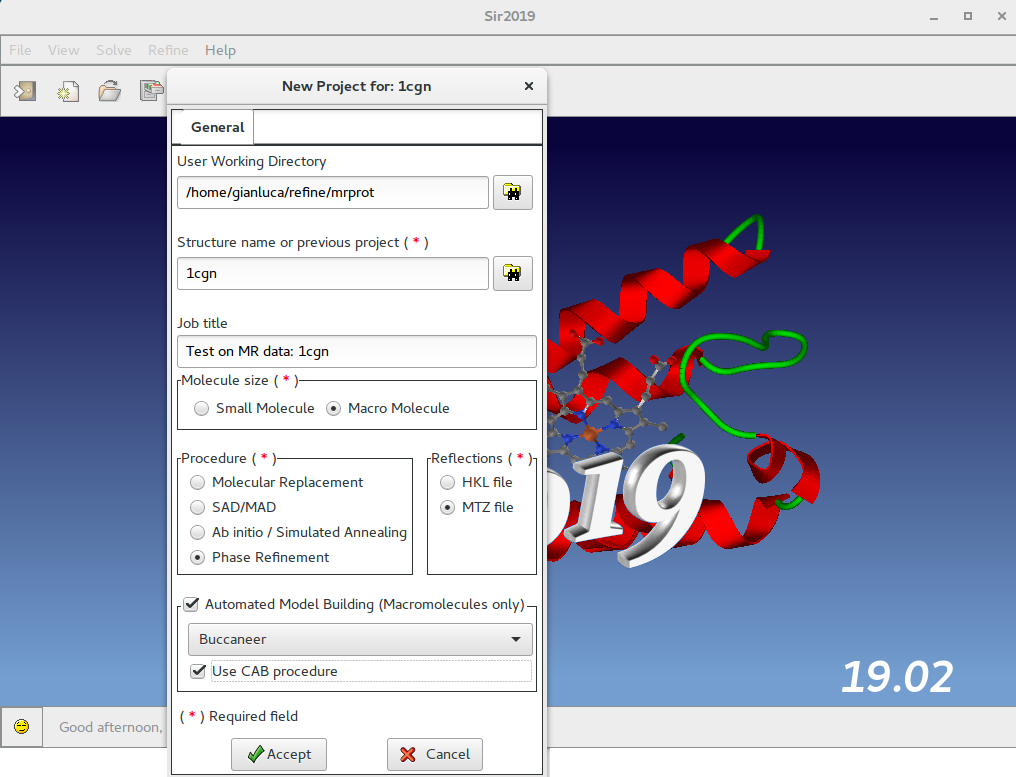
In this window the user should specify, among others, the working directory, the molecule size, the procedure to use (Phase Refinement) and the reflection file format (MTZ).
If an Automated Model Building program is installed, it is possible to use it by selecting Buccaneer or Nautilus or ARP/wARP or Phenix. If selected the CAB procedure will be applied at the end of this procedure.
Once clicked on ![]() it is possible to access the MTZ section:
it is possible to access the MTZ section:
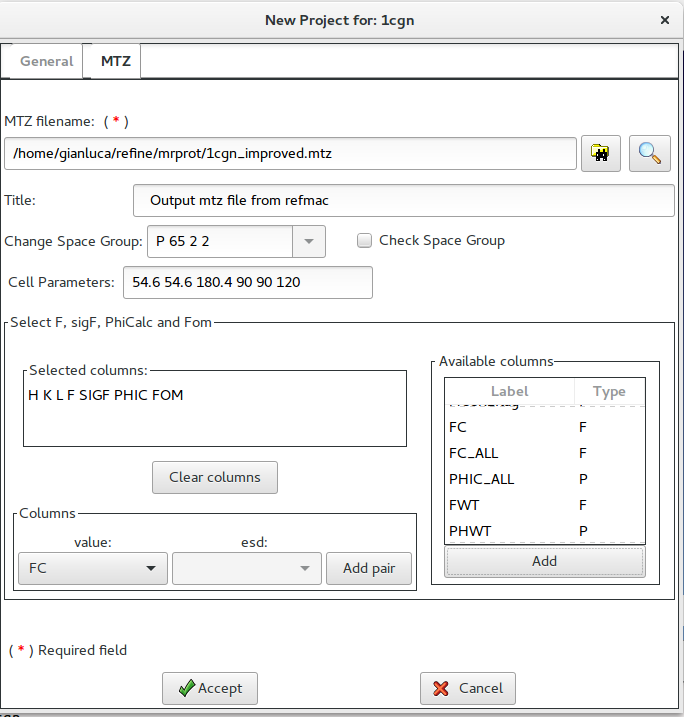
Using this window it is possible to supply the name of the MTZ file and to select the necessary columns (H K L F SigF, PhiCalc and Fom).
Once clicked on ![]() it is possible to access the Cell Content section:
it is possible to access the Cell Content section:
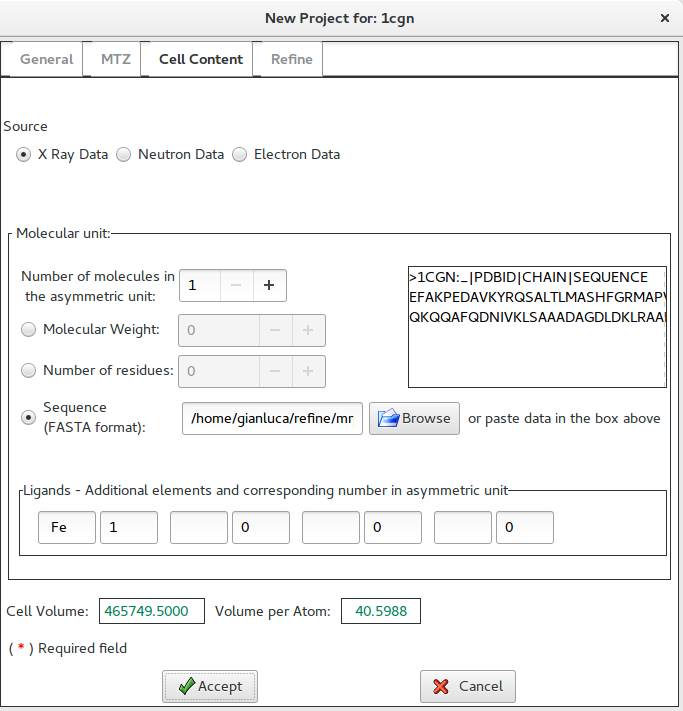
In this example, the sequence (in FASTA format) has been supplied.
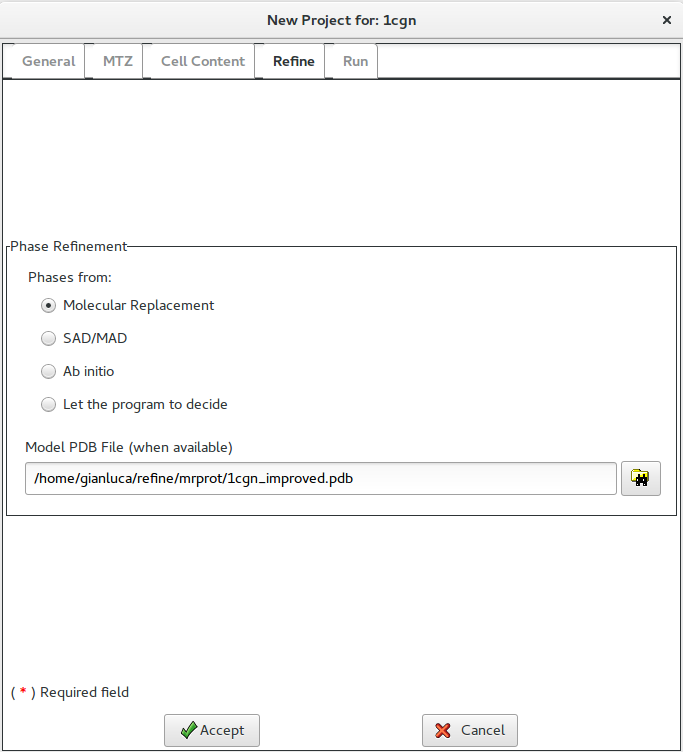
After data have been accepted, the Refine interface below is available, where the user may specify the method by which phases to refine were obtained. In case of Molecular Replacement a preliminary model may be supplied.
Once clicked on ![]() it is possible to access the Run section:
it is possible to access the Run section:
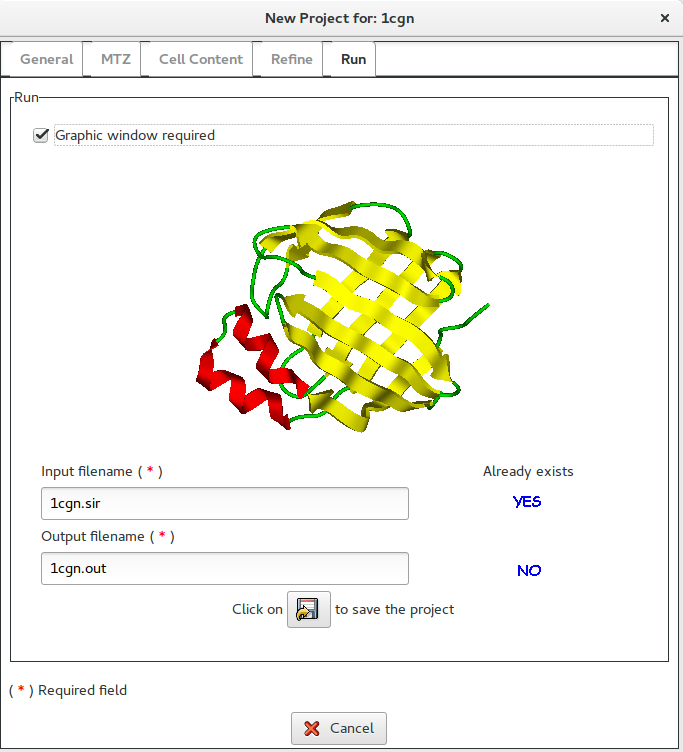
If Graphic window required is checked, a graphical output will be produced (see below).
In this section it is possible to change the default names for the input (project) and for the output file. The user, by clicking on ![]() button, can save the input file for Sir2019.
button, can save the input file for Sir2019.
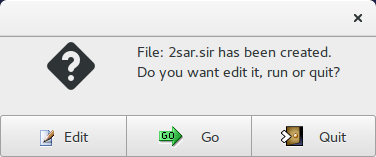
Now it is possible to edit ![]() the input file (2sar.sir); to start calculations click on
the input file (2sar.sir); to start calculations click on ![]() .
.
Once the program is over, by means of the “View output file” ![]() feature in “File” it is possible to access the complete output file.
feature in “File” it is possible to access the complete output file.
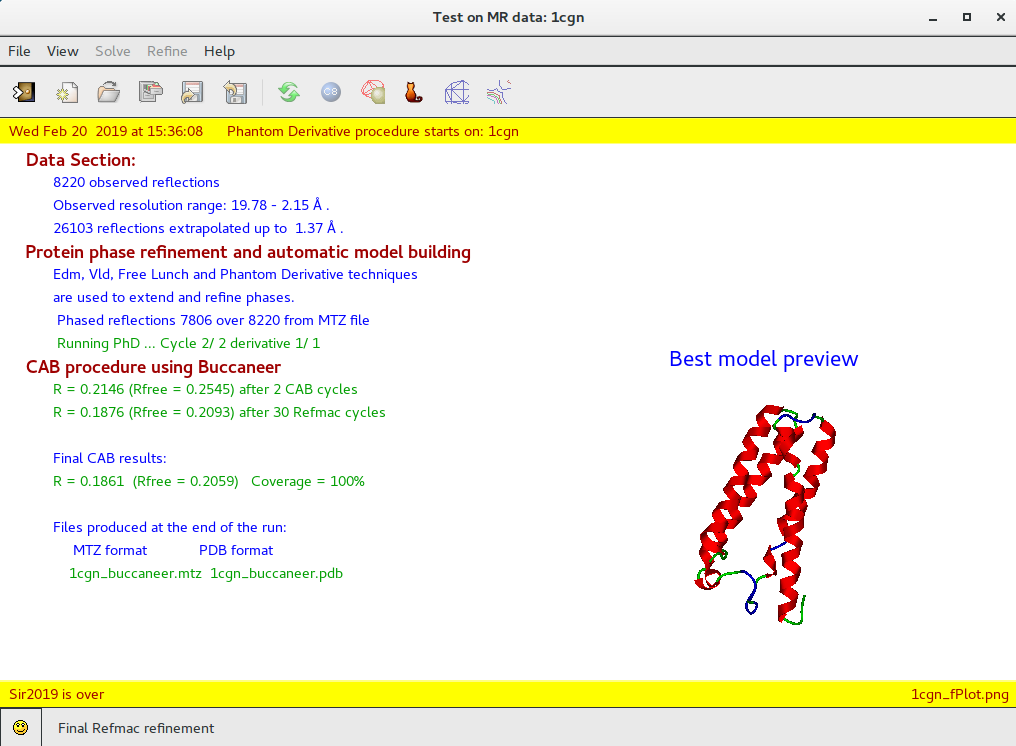
By means of the “Summary” ![]() feature in “File” menu is possible to get a synthetic output and also the graphic windows produced by the program during the run (in HTML format). An example of graphical output follows.
feature in “File” menu is possible to get a synthetic output and also the graphic windows produced by the program during the run (in HTML format). An example of graphical output follows.
When the program is over it is possible to interact with the structure using ![]() Jav:
Jav:
MODEL string
String is the name of the coordinates file (*.pdb) where a previously created model is stored . (i.e. a Molecular Replacement result).
MTZ string
String is the name of the reflections file in MTZ format, if not already defined in %Data. Phase and fom columns are necessary to perform refinement.
LABELS strings
Strings are the names of columns in the MTZ reflections file, if not already defined in %Data.
MR
Phases have been produced by a Molecular Replacement procedure.
MAD
Phases have been produced by a MAD/SAD procedure.
ABI
Phases have been produced by an ab initio procedure.